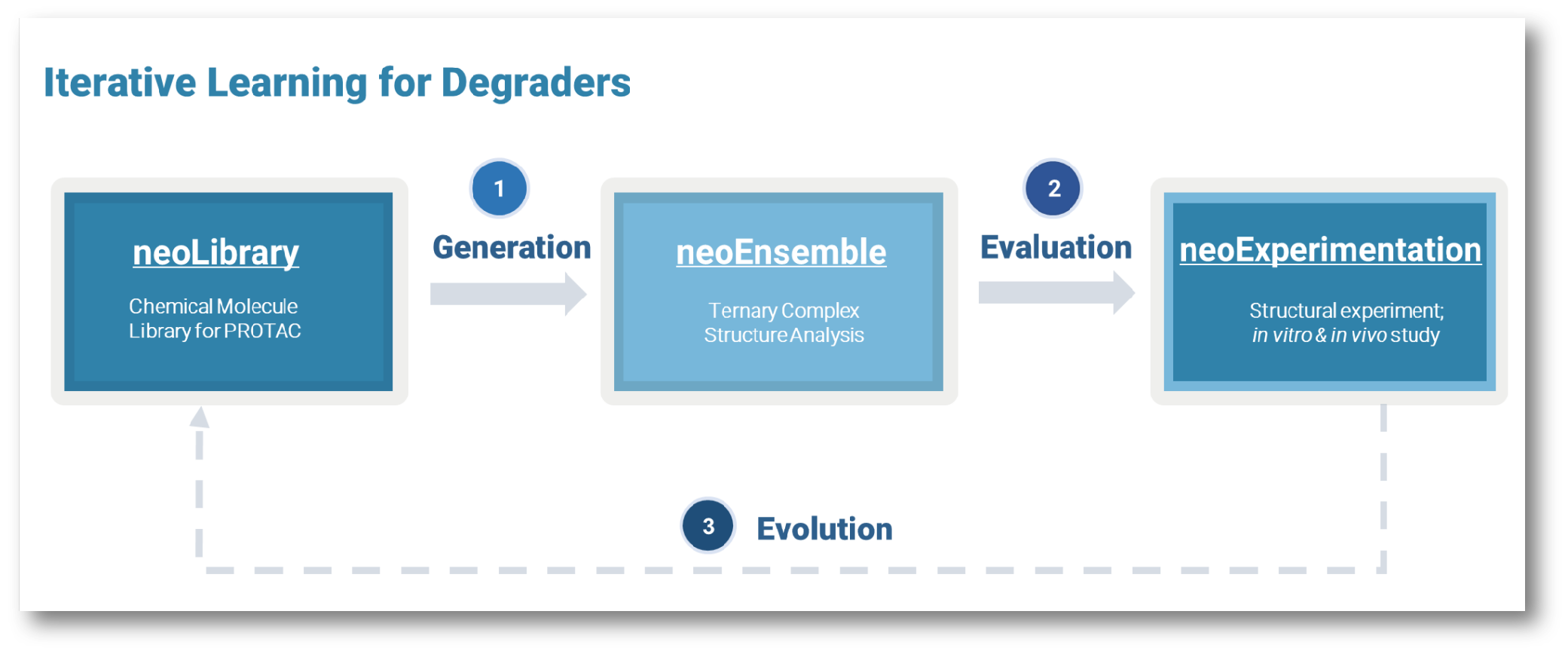
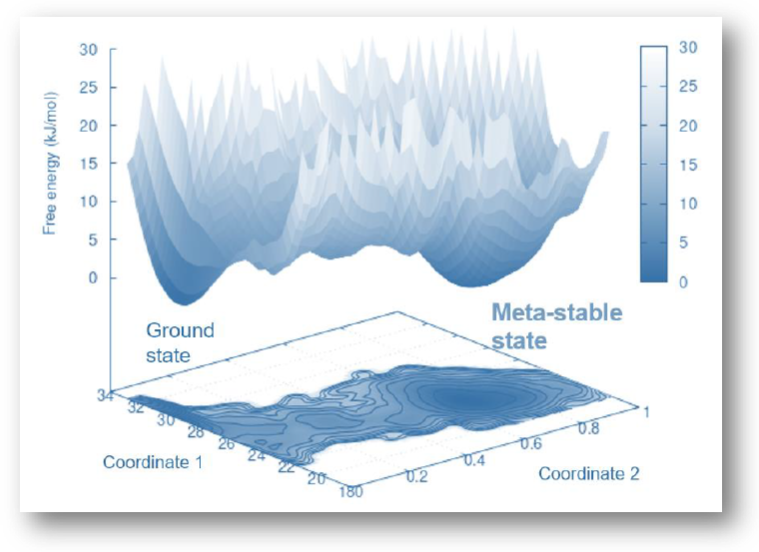
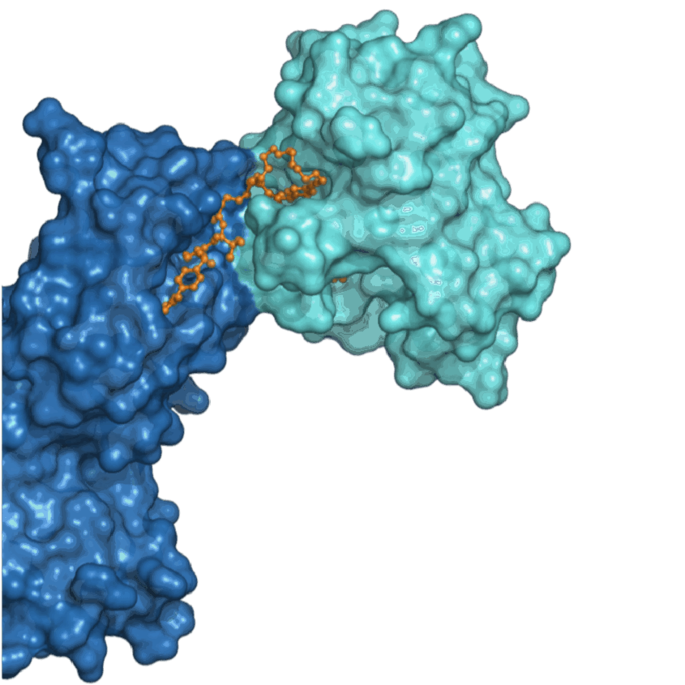
Dynamics/Motion based Protein-Protein Interface Design
Computational and experimental techniques are employed to locate regions on the
surface of proteins that can undergo conformational changes, potentially influencing
the binding affinity of protein-protein interactions. Moreover, in the design
process of biologics and degraders, function-associated metastable states of the
protein ensembles are also leveraged and captured, allowing for custom adaptability
to meet specific functional requirements of the interaction.
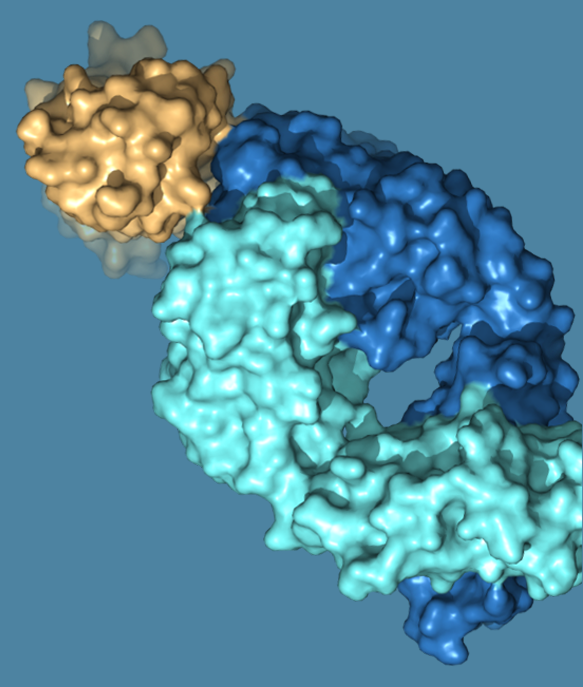

MetaPPI: neoBiologics™
AI-powered Synthetic Library for Drugs Discovery
For biological macromolecular drugs, traditional antibody expression display
platforms such as phage display and yeast display are insufficient to meet the
scanning of the entire antibody sequence based on library capacity restrictions, and
neoX Biotech originally performed virtual screening through calculation to establish
a smart library, narrow the screening region, and obtain feedback data through phage
or yeast display platforms for further optimization, and finally obtain antibodies
with better biological activity and druglikeness to achieve "Best-in-Class".

MetaPPI: neoDegrader™
Rational design for degraders by computation
Protein degraders are currently the focus of many pharmaceutical companies and are
considered to target undruggable targets. By designing degrader-induced
protein-protein interface and virtual screening of protein degradation compound
libraries, neoX Biotech can optimize the "target protein-protein degrader-E3 ligase"
ternary complexes to design lead compounds more efficiently and provide AI-optimized design and evaluations for drug discovery.